[Source: Eurosurveillance, full page: (LINK). Edited.]
Eurosurveillance, Volume 18, Issue 47, 21 November 2013
Research articles
Spatial and temporal analysis of human infection with avian influenza A(H7N9) virus in China, 2013
W Liu<SUP>1</SUP>, K Yang<SUP>2</SUP>, X Qi<SUP>1</SUP>, K Xu<SUP>1</SUP>, H Ji<SUP>1</SUP>, J Ai<SUP>1</SUP>, A Ge<SUP>1</SUP>, Y Wu<SUP>1</SUP>, Y Li<SUP>1</SUP>, Q Dai<SUP>1</SUP>, Q Liang<SUP>1</SUP>, C Bao<SUP>1</SUP>, R Bergquist<SUP>3</SUP>, F Tang<SUP>1</SUP>, Y Zhu <SUP>1</SUP>
<SUP></SUP>
<SUP>1</SUP>Department of Acute Infectious Disease Control and Prevention, Jiangsu Provincial Center for Disease Control and Prevention, Nanjing, China; 2 Department of Schistosomiasis Control, Jiangsu Institute of Parasitic Diseases, Wuxi, China; 3 Ingerod 407, Brastad, Sweden
________
Citation style for this article: Liu W, Yang K, Qi X, Xu K, Ji H, Ai J, Ge A, Wu Y, Li Y, Dai Q, Liang Q, Bao C, Bergquist R, Tang F, Zhu Y. Spatial and temporal analysis of human infection with avian influenza A(H7N9) virus in China, 2013. Euro Surveill. 2013;18(47):pii=20640. Available online: http://www.eurosurveillance.org/View...rticleId=20640
Date of submission: 22 July 2013
________
Descriptive and geographic information system methods were used to depict the spatial and temporal characteristics of the outbreak of human infection with a novel avian influenza A(H7N9) virus in mainland China, the peak of which appeared between 28 March and 18 April 2013. As of 31 May 2013, there was a total of 131 reported human infections in China, with a cumulative mortality of 29% (38/131). The outbreak affected 10 provinces, with 106 of the cases being concentrated in the eastern coastal provinces of Zhejiang, Shanghai and Jiangsu. Statistically significant spatial clustering of cumulative human cases was identified by the Cuzick?Edwards? k-nearest neighbour method. Three spatio-temporal clusters of cases were detected by space?time scan analysis. The principal cluster covered 18 counties in Zhejiang during 3 to 18 April (relative risk (RR): 26.39;p<0.0001), while two secondary clusters in March and April covered 21 counties along the provincial boundary between Shanghai and Jiangsu (RR: 6.35;p<0.0001) and two counties in Jiangsu (RR: 72.48;p=0.0025). The peak of the outbreak was in the eastern coastal provinces of Zhejiang, Shanghai and Jiangsu that was characterised by statistically significant spatio-temporal aggregation, with a particularly high incidence in March and April 2013.
_________
Introduction
A novel avian influenza A(H7N9) virus emerged in China in February 2013, causing an outbreak of human infections characterised by rapidly progressive severe illness and fatal outcome [1,2]. After the first case of human infection was confirmed in the Minhang district of Shanghai on 31 March 2013, the disease gradually spread to the three neighbouring provinces of Zhejiang, Jiangsu and Anhui during the following four weeks.
The number of cases was very low in this period but then began to increase rapidly, which raised serious concern throughout the world [3]. The Chinese government responded swiftly and the containment measures applied (shutting down live poultry markets) effectively controlled the outbreak ? an approach strongly commended by the international community [4,5]. To date, the influenza A(H7N9) virus has not been detected in people or birds outside China.
Currently, the outbreak has entered in a stationary state in China and new infections appear only rarely [6]. However, scientists and authorities are still concerned that there might be a risk of a pandemic developing [6].
Strong efforts to date have been made to understand this new virus and substantial progress in epidemiology, diagnosis, therapy and aetiology of the infection has been achieved [1,7-12]. However, much remains to be discovered [13], particularly with regard to the spatial and temporal pattern of this emerging infectious disease. Understanding such patterns is essential for effective surveillance and control. A sound description of the dynamic progress of the outbreak is needed to refine control strategies and optimise resource allocation.
Advances in geographic information system (GIS) and related technologies have fostered new opportunities for examining diseases. As a distinct, useful tool for spatio-temporal analysis, GIS has been widely applied in controlling emerging infectious diseases, such as severe acute respiratory syndrome (SARS) [14,15], avian influenza A(H5N1) [16] and influenza A(H1N1)pdm2009 [17].
In the present study, we concurrently used descriptive analysis and GIS methods to depict the spatial and temporal characteristics of the outbreak of human infection with influenza A(H7N9) virus in mainland China. This work was conducted with the expectation that the results might provide some clues for further epidemiological research and help scientists and health authorities to more effectively target and improve their surveillance and control efforts in the future.
Methods
Case definition
The case definition was based on The diagnosis and treatment programs of human infection with influenza A(H7N9) virus[18] issued by National Health and Family Planning Commission of the People?s Republic of China, in which a confirmed case of human infection with influenza A(H7N9) virus is defined as a patient with influenza-like illness, or a suspected case with respiratory specimens, testing positive for the influenza A(H7N9) virus by one of the following laboratory diagnostic tests: isolation of influenza A(H7N9) virus or real-time reverse transcription polymerase chain reaction (rRT-PCR) assay for influenza A(H7N9) virus.
A suspected case is defined as a person presenting with unexplained acute lower respiratory illness with fever (≥38 ?C) and cough, shortness of breath or difficulty breathing or infiltrates or evidence of an acute pneumonia on chest radiograph plus evidence of respiratory failure (hypoxemia, severe tachypnoea), and either (i) positive laboratory confirmation of an influenza A virus infection but insufficient laboratory evidence for influenza A(H7N9) virus infection because of lack of specimens or (ii) epidemiologically linked to a confirmed influenza A(H7N9) case, but without any respiratory specimens available for influenza A(H7N9) virus testing [18].
Data source
As of 31 May 2013, a total of 131 confirmed human infections with influenza A(H7N9) virus including 38 deaths had occurred in mainland China. For each case, we collected the following data: sex, age, place of residence (postal address), the date of symptom onset and, in case of fatal outcome, the date of death. This information was extracted from the influenza A(H7N9) outbreak reporting released by National Health and Family Planning Commission of the People?s Republic of China. We obtained population data from the Scientific Data Sharing Center of Public Health [19], developed by Chinese Center for Disease Control and Prevention. The maps were downloaded from the National Geomatics Center of China [20].
Study areas and study period
A total of 10 provinces in east China, representing more than a million square kilometres and approximately 570 million inhabitants (Figure 1) reported cases of influenza A(H7N9) virus infection. The study period was from 19 February to 31 May 2013.
_____
Figure 1. Population densities in mainland China (A)<SUP>a</SUP> and spatio-temporal cluster analysis of human cases of influenza A(H7N9) virus infection in three eastern provinces, China (B and C)
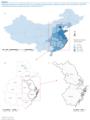
______
Statistical analysis
Descriptive statistics were used to illustrate the characteristics of the population distribution and temporal distribution of the cases of human influenza A(H7N9) virus infection and related deaths. Pearson correlation analysis was conducted to examine the presumed association between the incidence of human infection and population density at the county level using SAS software version 8.1 (SAS Institute Inc., Cary, NC, United States).
Spatial autocorrelation analysis
Spatial autocorrelation provides an analysis of the degree of the dependency among various observations in geographical space [21]. In this study, the Cuzick?Edwards? k-nearest neighbour method was used to identify the possibility of spatial clustering [22]. Existence of potential spatial clustering was analysed at each of the first 10 neighbourhood levels, and the overall p value was adjusted for multiple comparisons with the Simes approach based on ClusterSeer software version 2.5.1 (BioMedware, Ann Arbor, MI, United States).
Space?time scan statistic
SaTScan software version 9.0 [23] was employed to conduct a retrospective space?time scan statistic to identify whether or not the human influenza A(H7N9) virus infections were randomly distributed over space and time in Zhejiang, Shanghai and Jiangsu, the three eastern coastal provinces with the highest number of cases. The method is based on the use of a cylindrical window, which represents the circular geographical base of a cylinder, the height of which corresponds to time. The null hypothesis assumes that the relative risk (RR) of human infections is the same within the window compared with outside it. In this study, the human influenza A(H7N9) virus infections were assumed as having a Poisson distribution in each location. The spatial size of scan window was limited to 25% of the total population and the length of time was limited to half of the whole study period. The statistical significance of each cluster was based on comparing the log likelihood ratio (LLR) against a null distribution obtained through Monte Carlo simulation, with the number of replications set to 999 and the significance level set as 0.05. The window with the maximum LLR was deemed the cluster least likely to be due to chance. Other windows with a statistically significant LLR were considered as secondary clusters [24,25].
Results
Demographic characteristics of cases
During the study period, a total of 131 confirmed human cases were reported, 93 of which were male (71%) and 38 female (29%). The age of the cases ranged from two to 91 years, with a median of 61 years. Male cases aged 60 years or older accounted for 39% (51/131) of all cases. Of the 131 cases, 38 (29%) had died from the infection. Full data were not available for one case, but of the remaining 37 fatal cases, 29 were male and 8 female. Their ages ranged from 27 to 91 years, with a median of 67 years, with 28 aged 60 years or older. The sex-related mortality was 31% (29/93) for the male cases and 21% (8/38) for the female cases. No statistically significant difference was observed between the two groups (chi-square test statistic: 1.3660;p=0.2425). The interval between the date of symptom onset and the date of death ranged from six to 67 days, with a median of 20 days.
Temporal characteristics of cases
We divided the study period into 15 weeks of seven days (with week 1 starting 19 February 2013) except the last week, which included only four days (i.e. 28?31 May 2013) and calculated the number of cases for each week (Figure 2). The first two cases developed symptoms simultaneously on 19 February. The outbreak was highly sporadic in the four weeks after the first cases developed symptoms, with just 11 sporadic cases, after which the number of cases increased substantially. Weeks 5 to 9 (19 March?22 April) included the great majority of cases (n=113), with most of the mortality occurring during weeks 7 to 12 (2 April?13 May). There was a peak of incidence between 28 March and 18 April, with76% (100/131) of all cases. From week 10, the incidence decreased rapidly. Only two cases occurred in May, the last month of the study period.
________
Figure 2. Daily (A) and weekly<SUP>a</SUP> (B) distribution of human cases of influenza A(H7N9) virus infection (n=131) and related deaths (n=38), China, 19 February?31 May 2013

________
Spatial characteristics
In the study period, 10 provinces successively reported human infections of influenza A(H7N9) virus. More than 80% of cases (106/131) occurred in Zhejiang, Shanghai and Jiangsu, the three eastern coastal provinces. At the county level, just 70 counties had ever reported cases, accounting for approximately 7% (70/971) of the total number of counties in the 10 provinces, most of which are located along the provincial boundaries among Zhejiang, Jiangsu and Shanghai. Of these 70 counties, 43 had only one case, while five had five or more (Figure 3). Although a relatively high population density was observed in the regions with human infections, the Pearson correlation analysis (r: 0.17;p=0.1702) suggested that there was no statistically significant association between the incidence and population density. The Cuzick?Edwards? test identified statistically significant spatial clustering of cases, not only for each level of neighbourhoods, but also for a summary test at the significance level of 0.05, implying that the distribution of human cases was spatially autocorrelated.
_________
Figure 3. Population density at county level<SUP>a</SUP> (A) and spatial distribution of human cases of influenza A(H7N9) virus infection (n=131) (B) in 10 eastern provinces, China, 19 February?31 May 2013

________
Dynamic progress of the outbreak
Cases of confirmed influenza A(H7N9) virus infection were initially very rare in weeks 1?4 (19 February?18 March). The outbreak started in Shanghai and spread to Zhejiang, Jiangsu, and Anhui in this period. A period of high incidence appeared subsequently between weeks 5 and 9 (19 March?22 April). In this stage, the outbreak gradually spread to another six provinces: Henan, Beijing, Hunan, Jiangxi, Shandong and Fujian, in that order. But Zhejiang, Jiangsu and Shanghai were the most affected areas, with 87 cases. After that, the outbreak decreased rapidly. In the last six weeks of the study period, only seven new infections occurred and the outbreak did not spread to any other provinces. Notably, no infections occurred during this period in the highly endemic regions of Zhejiang, Shanghai and Jiangsu. No infections occurred from weeks 12 to 13 (7?20 May) throughout the study area. For details, see Figure 4.
_________
Figure 4. Spatial diffusion of human cases of influenza A(H7N9) virus infection in 10 eastern provinces, China, 19 February?31 May 2013 (n=131)

________
Spatio-temporal cluster analysis
Figure 5 shows the results of the space?time scan statistic analysis in the three eastern coastal provinces, which had the highest number of human cases. Three clusters were detected: one principal and two secondary. The principal cluster (LLR:77.72) had a RR of 26.39: it included 37 cases and covered 18 counties in Zhejiang, lasting from 3 to 18 April. One of the secondary clusters, with an LLR of 25.06, included 21 counties (17 in Shanghai and four in Jiangsu). With 28 cases, it lasted from 25 March to 11 April and had a RR of 6.35. The other secondary cluster (LLR:16.37) covered just two counties in Jiangsu: it lasted from 4 to 8 April, had five cases, with a RR of 72.48.
________
Figure 5. Spatio-temporal clusters of human cases of influenza A(H7N9) virus infection at county level in Zhejiang, Shanghai and Jiangsu provinces, China, 19 February?31 May 2013 (n=106)
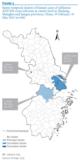
________
Discussion
The outbreak of human infection with influenza A(H7N9) virus has attracted considerable attention because it is an emerging infectious disease caused by a novel, reassortant avian-origin influenza virus [26,27]. As this virus does not have the ability to be transmitted from person to person, it is unlikely to lead to a pandemic, but gene sequences analysis has indicated that it is better adapted than other avian influenza viruses to infect mammals [9,28]. Scientists are therefore concerned about the possibility of a pandemic if there is further gene mutation [29,30].
In the study period, the outbreak caused 131 confirmed human infections in mainland China. As reported previously [2], most cases were male, mainly aged 60 years or older. In contrast, the influenza A(H5N1) virus mainly affected children, adolescents and young adults (aged between 4 and 29 years, with a median of 18 years) [31]. As of 31 May, 38 cases of influenza A(H7N9) virus infection had died, with accumulative mortality of 29%, much higher than the 21% (17/82) in a previous report on the outbreak [2]. In comparison with the influenza A(H1N1)pdm2009 virus infections (mortality: 2.5%), the mortality of cases of influenza A(H7N9) virus infection was higher[32], although it was clearly lower than that of cases of highly pathogenic avian influenza A(H5N1) virus infection (mortality: 56%) [31]. As shown in a former case study [2], most of the human cases of influenza A(H7N9) virus infection had a severe illness (77/82), while others manifested as mild illness (4/82). In addition, a four year-old child in Beijing was identified as an asymptomatic carrier of influenza A(H7N9) virus [33]. This has been the only asymptomatic infection of influenza A(H7N9) virus to date. Thus, human infections with influenza A(H7N9) virus may result in fatal disease, mild illness as well as silent infection.
Our study shows that the influenza A(H7N9) outbreak in mainland China can be divided into 3 stages: early (weeks1?4), high-incidence (weeks5?9) and late stationary (weeks10?15). In the early stage, the outbreak was considered as non-threatening and highly sporadic. However, the high-incidence stage (in March?April) represented a peak, with 76% (100/131) of the cases during this stage. Furthermore, the three spatio-temporal clusters of cases identified were all during this period. Increased surveillance and control should be carried out in this period in future, to find out if there is an association with climate. In the late stationary stage, transmission was effectively controlled, leading to the ending of the outbreak in the most severely affected areas. This is now believed to be mainly due to the closure of live poultry markets [5].
Some studies have claimed that climatic factors, particularly temperature, have a clear impact on seasonal influenza outbreaks [34,35]. The weather in eastern China is very cold between December and January, and temperatures begin to rise in late February. It is usually relatively warm in March and April, and starts to get hot from May. The temporal characteristics of human infections with influenza A(H7N9) virus suggest that temperature has some association with incidence of this disease. The prevailing mild climate may have been particularly suitable for influenza A(H7N9) virus infection since there was an increase in the number of cases in March?April. If there is a relationship, attention must be paid as there could be a potential outbreak in the same period (March?April) in the future.
The outbreak was limited to 10 provinces in eastern China. In-depth studies of the factors driving the spatial diffusion of cases are essential to prevent future outbreaks. It is notable that when the first human infections appeared in Beijing and Henan, there were no cases in the neighbouring provinces. What is the reason for this? Were the two regions new, natural foci of the influenza A(H7N9) virus strain? Alternatively, and perhaps more probable, could they be attributed to the regional poultry trade or migration of infected wild birds? These possibilities need to be further studied.
The three coastal provinces of Zhejiang, Shanghai and Jiangsu were the most severely affected areas. Understanding the main factors causing this spatial concentration is critical in the approach to control this type of emerging disease. Some studies report that the spatial distribution of human infection with highly pathogenic avian influenza A(H5N1) is mainly associated with human population density, poultry density, elevation and the proportion of land covered by water bodies [16,36]. However, our study suggests that population density does not influence the spatial pattern of human infection with influenza A(H7N9) virus. Further studies are needed to confirm whether or not there are other counterintuitive correlations involved in the spatial distribution of human infection due to this virus.
The SatScan method has become an increasingly popular adjunct for exploring the spatio-temporal distribution of infectious diseases [37]. We selected this method in the hope of identifying clustering of cases and found three clusters in the three coastal provinces with a high incidence, which were proposed to be the most probable place of the viral reassortment [12,38]. Two of the identified clusters represent the most important hotspots of the outbreak in China and were found in the counties along the boundaries between these three provinces. Surveillance should be focused on these regions to identify any future emergence of this or other new virus strains and monitor possible further mutation or reassortment that may alter the risks to animals or humans. The third cluster covered just two counties within Jiangsu province and was situated far from the other two clusters. In addition, its duration was very short. The risk factors that contribute to clustering warrant further study.
This study, using spatio-temporal methods in conjunction with classic descriptive analysis, found evidence of clustering, in both space and time. The results can provide clues for further developments and serve as a reference and basis for the surveillance and control of human infection with influenza A(H7N9) virus in the future.
_____
Acknowledgements
This work was supported by the Jiangsu Province Health Development Project with Science and Education (ZX201109, RC2011085 and RC 2011094), and the research project of Jiangsu preventive medicine (NO.YZ201020).
Conflict of interest
None declared.
Authors? contributions
Wendong Liu and Kun Yang contributed equally to this study. Correspondence to Yefei Zhu and Fenyang Tang. Yefei Zhu and Fenyang Tang designed the study; Wendong Liu, Kun Yang, Xian Qi, KeXu, Hong Ji, Jing Ai, Aihua Ge, Ying Wu, Yuan Li, Qigang Dai, Qi Liang and Changjun Bao collected, analyzed and interpreted data; Wendong Liu, Kun Yang, and Yefei Zhu drafted the article; Robert Bergquist revised the manuscript. All authors reviewed and revised the first and final drafts of this manuscript.
References
-
--------
Eurosurveillance, Volume 18, Issue 47, 21 November 2013
Research articles
Spatial and temporal analysis of human infection with avian influenza A(H7N9) virus in China, 2013
W Liu<SUP>1</SUP>, K Yang<SUP>2</SUP>, X Qi<SUP>1</SUP>, K Xu<SUP>1</SUP>, H Ji<SUP>1</SUP>, J Ai<SUP>1</SUP>, A Ge<SUP>1</SUP>, Y Wu<SUP>1</SUP>, Y Li<SUP>1</SUP>, Q Dai<SUP>1</SUP>, Q Liang<SUP>1</SUP>, C Bao<SUP>1</SUP>, R Bergquist<SUP>3</SUP>, F Tang<SUP>1</SUP>, Y Zhu <SUP>1</SUP>
<SUP></SUP>
<SUP>1</SUP>Department of Acute Infectious Disease Control and Prevention, Jiangsu Provincial Center for Disease Control and Prevention, Nanjing, China; 2 Department of Schistosomiasis Control, Jiangsu Institute of Parasitic Diseases, Wuxi, China; 3 Ingerod 407, Brastad, Sweden
________
Citation style for this article: Liu W, Yang K, Qi X, Xu K, Ji H, Ai J, Ge A, Wu Y, Li Y, Dai Q, Liang Q, Bao C, Bergquist R, Tang F, Zhu Y. Spatial and temporal analysis of human infection with avian influenza A(H7N9) virus in China, 2013. Euro Surveill. 2013;18(47):pii=20640. Available online: http://www.eurosurveillance.org/View...rticleId=20640
Date of submission: 22 July 2013
________
Descriptive and geographic information system methods were used to depict the spatial and temporal characteristics of the outbreak of human infection with a novel avian influenza A(H7N9) virus in mainland China, the peak of which appeared between 28 March and 18 April 2013. As of 31 May 2013, there was a total of 131 reported human infections in China, with a cumulative mortality of 29% (38/131). The outbreak affected 10 provinces, with 106 of the cases being concentrated in the eastern coastal provinces of Zhejiang, Shanghai and Jiangsu. Statistically significant spatial clustering of cumulative human cases was identified by the Cuzick?Edwards? k-nearest neighbour method. Three spatio-temporal clusters of cases were detected by space?time scan analysis. The principal cluster covered 18 counties in Zhejiang during 3 to 18 April (relative risk (RR): 26.39;p<0.0001), while two secondary clusters in March and April covered 21 counties along the provincial boundary between Shanghai and Jiangsu (RR: 6.35;p<0.0001) and two counties in Jiangsu (RR: 72.48;p=0.0025). The peak of the outbreak was in the eastern coastal provinces of Zhejiang, Shanghai and Jiangsu that was characterised by statistically significant spatio-temporal aggregation, with a particularly high incidence in March and April 2013.
_________
Introduction
A novel avian influenza A(H7N9) virus emerged in China in February 2013, causing an outbreak of human infections characterised by rapidly progressive severe illness and fatal outcome [1,2]. After the first case of human infection was confirmed in the Minhang district of Shanghai on 31 March 2013, the disease gradually spread to the three neighbouring provinces of Zhejiang, Jiangsu and Anhui during the following four weeks.
The number of cases was very low in this period but then began to increase rapidly, which raised serious concern throughout the world [3]. The Chinese government responded swiftly and the containment measures applied (shutting down live poultry markets) effectively controlled the outbreak ? an approach strongly commended by the international community [4,5]. To date, the influenza A(H7N9) virus has not been detected in people or birds outside China.
Currently, the outbreak has entered in a stationary state in China and new infections appear only rarely [6]. However, scientists and authorities are still concerned that there might be a risk of a pandemic developing [6].
Strong efforts to date have been made to understand this new virus and substantial progress in epidemiology, diagnosis, therapy and aetiology of the infection has been achieved [1,7-12]. However, much remains to be discovered [13], particularly with regard to the spatial and temporal pattern of this emerging infectious disease. Understanding such patterns is essential for effective surveillance and control. A sound description of the dynamic progress of the outbreak is needed to refine control strategies and optimise resource allocation.
Advances in geographic information system (GIS) and related technologies have fostered new opportunities for examining diseases. As a distinct, useful tool for spatio-temporal analysis, GIS has been widely applied in controlling emerging infectious diseases, such as severe acute respiratory syndrome (SARS) [14,15], avian influenza A(H5N1) [16] and influenza A(H1N1)pdm2009 [17].
In the present study, we concurrently used descriptive analysis and GIS methods to depict the spatial and temporal characteristics of the outbreak of human infection with influenza A(H7N9) virus in mainland China. This work was conducted with the expectation that the results might provide some clues for further epidemiological research and help scientists and health authorities to more effectively target and improve their surveillance and control efforts in the future.
Methods
Case definition
The case definition was based on The diagnosis and treatment programs of human infection with influenza A(H7N9) virus[18] issued by National Health and Family Planning Commission of the People?s Republic of China, in which a confirmed case of human infection with influenza A(H7N9) virus is defined as a patient with influenza-like illness, or a suspected case with respiratory specimens, testing positive for the influenza A(H7N9) virus by one of the following laboratory diagnostic tests: isolation of influenza A(H7N9) virus or real-time reverse transcription polymerase chain reaction (rRT-PCR) assay for influenza A(H7N9) virus.
A suspected case is defined as a person presenting with unexplained acute lower respiratory illness with fever (≥38 ?C) and cough, shortness of breath or difficulty breathing or infiltrates or evidence of an acute pneumonia on chest radiograph plus evidence of respiratory failure (hypoxemia, severe tachypnoea), and either (i) positive laboratory confirmation of an influenza A virus infection but insufficient laboratory evidence for influenza A(H7N9) virus infection because of lack of specimens or (ii) epidemiologically linked to a confirmed influenza A(H7N9) case, but without any respiratory specimens available for influenza A(H7N9) virus testing [18].
Data source
As of 31 May 2013, a total of 131 confirmed human infections with influenza A(H7N9) virus including 38 deaths had occurred in mainland China. For each case, we collected the following data: sex, age, place of residence (postal address), the date of symptom onset and, in case of fatal outcome, the date of death. This information was extracted from the influenza A(H7N9) outbreak reporting released by National Health and Family Planning Commission of the People?s Republic of China. We obtained population data from the Scientific Data Sharing Center of Public Health [19], developed by Chinese Center for Disease Control and Prevention. The maps were downloaded from the National Geomatics Center of China [20].
Study areas and study period
A total of 10 provinces in east China, representing more than a million square kilometres and approximately 570 million inhabitants (Figure 1) reported cases of influenza A(H7N9) virus infection. The study period was from 19 February to 31 May 2013.
_____
Figure 1. Population densities in mainland China (A)<SUP>a</SUP> and spatio-temporal cluster analysis of human cases of influenza A(H7N9) virus infection in three eastern provinces, China (B and C)

______
Statistical analysis
Descriptive statistics were used to illustrate the characteristics of the population distribution and temporal distribution of the cases of human influenza A(H7N9) virus infection and related deaths. Pearson correlation analysis was conducted to examine the presumed association between the incidence of human infection and population density at the county level using SAS software version 8.1 (SAS Institute Inc., Cary, NC, United States).
Spatial autocorrelation analysis
Spatial autocorrelation provides an analysis of the degree of the dependency among various observations in geographical space [21]. In this study, the Cuzick?Edwards? k-nearest neighbour method was used to identify the possibility of spatial clustering [22]. Existence of potential spatial clustering was analysed at each of the first 10 neighbourhood levels, and the overall p value was adjusted for multiple comparisons with the Simes approach based on ClusterSeer software version 2.5.1 (BioMedware, Ann Arbor, MI, United States).
Space?time scan statistic
SaTScan software version 9.0 [23] was employed to conduct a retrospective space?time scan statistic to identify whether or not the human influenza A(H7N9) virus infections were randomly distributed over space and time in Zhejiang, Shanghai and Jiangsu, the three eastern coastal provinces with the highest number of cases. The method is based on the use of a cylindrical window, which represents the circular geographical base of a cylinder, the height of which corresponds to time. The null hypothesis assumes that the relative risk (RR) of human infections is the same within the window compared with outside it. In this study, the human influenza A(H7N9) virus infections were assumed as having a Poisson distribution in each location. The spatial size of scan window was limited to 25% of the total population and the length of time was limited to half of the whole study period. The statistical significance of each cluster was based on comparing the log likelihood ratio (LLR) against a null distribution obtained through Monte Carlo simulation, with the number of replications set to 999 and the significance level set as 0.05. The window with the maximum LLR was deemed the cluster least likely to be due to chance. Other windows with a statistically significant LLR were considered as secondary clusters [24,25].
Results
Demographic characteristics of cases
During the study period, a total of 131 confirmed human cases were reported, 93 of which were male (71%) and 38 female (29%). The age of the cases ranged from two to 91 years, with a median of 61 years. Male cases aged 60 years or older accounted for 39% (51/131) of all cases. Of the 131 cases, 38 (29%) had died from the infection. Full data were not available for one case, but of the remaining 37 fatal cases, 29 were male and 8 female. Their ages ranged from 27 to 91 years, with a median of 67 years, with 28 aged 60 years or older. The sex-related mortality was 31% (29/93) for the male cases and 21% (8/38) for the female cases. No statistically significant difference was observed between the two groups (chi-square test statistic: 1.3660;p=0.2425). The interval between the date of symptom onset and the date of death ranged from six to 67 days, with a median of 20 days.
Temporal characteristics of cases
We divided the study period into 15 weeks of seven days (with week 1 starting 19 February 2013) except the last week, which included only four days (i.e. 28?31 May 2013) and calculated the number of cases for each week (Figure 2). The first two cases developed symptoms simultaneously on 19 February. The outbreak was highly sporadic in the four weeks after the first cases developed symptoms, with just 11 sporadic cases, after which the number of cases increased substantially. Weeks 5 to 9 (19 March?22 April) included the great majority of cases (n=113), with most of the mortality occurring during weeks 7 to 12 (2 April?13 May). There was a peak of incidence between 28 March and 18 April, with76% (100/131) of all cases. From week 10, the incidence decreased rapidly. Only two cases occurred in May, the last month of the study period.
________
Figure 2. Daily (A) and weekly<SUP>a</SUP> (B) distribution of human cases of influenza A(H7N9) virus infection (n=131) and related deaths (n=38), China, 19 February?31 May 2013

________
Spatial characteristics
In the study period, 10 provinces successively reported human infections of influenza A(H7N9) virus. More than 80% of cases (106/131) occurred in Zhejiang, Shanghai and Jiangsu, the three eastern coastal provinces. At the county level, just 70 counties had ever reported cases, accounting for approximately 7% (70/971) of the total number of counties in the 10 provinces, most of which are located along the provincial boundaries among Zhejiang, Jiangsu and Shanghai. Of these 70 counties, 43 had only one case, while five had five or more (Figure 3). Although a relatively high population density was observed in the regions with human infections, the Pearson correlation analysis (r: 0.17;p=0.1702) suggested that there was no statistically significant association between the incidence and population density. The Cuzick?Edwards? test identified statistically significant spatial clustering of cases, not only for each level of neighbourhoods, but also for a summary test at the significance level of 0.05, implying that the distribution of human cases was spatially autocorrelated.
_________
Figure 3. Population density at county level<SUP>a</SUP> (A) and spatial distribution of human cases of influenza A(H7N9) virus infection (n=131) (B) in 10 eastern provinces, China, 19 February?31 May 2013

________
Dynamic progress of the outbreak
Cases of confirmed influenza A(H7N9) virus infection were initially very rare in weeks 1?4 (19 February?18 March). The outbreak started in Shanghai and spread to Zhejiang, Jiangsu, and Anhui in this period. A period of high incidence appeared subsequently between weeks 5 and 9 (19 March?22 April). In this stage, the outbreak gradually spread to another six provinces: Henan, Beijing, Hunan, Jiangxi, Shandong and Fujian, in that order. But Zhejiang, Jiangsu and Shanghai were the most affected areas, with 87 cases. After that, the outbreak decreased rapidly. In the last six weeks of the study period, only seven new infections occurred and the outbreak did not spread to any other provinces. Notably, no infections occurred during this period in the highly endemic regions of Zhejiang, Shanghai and Jiangsu. No infections occurred from weeks 12 to 13 (7?20 May) throughout the study area. For details, see Figure 4.
_________
Figure 4. Spatial diffusion of human cases of influenza A(H7N9) virus infection in 10 eastern provinces, China, 19 February?31 May 2013 (n=131)

________
Spatio-temporal cluster analysis
Figure 5 shows the results of the space?time scan statistic analysis in the three eastern coastal provinces, which had the highest number of human cases. Three clusters were detected: one principal and two secondary. The principal cluster (LLR:77.72) had a RR of 26.39: it included 37 cases and covered 18 counties in Zhejiang, lasting from 3 to 18 April. One of the secondary clusters, with an LLR of 25.06, included 21 counties (17 in Shanghai and four in Jiangsu). With 28 cases, it lasted from 25 March to 11 April and had a RR of 6.35. The other secondary cluster (LLR:16.37) covered just two counties in Jiangsu: it lasted from 4 to 8 April, had five cases, with a RR of 72.48.
________
Figure 5. Spatio-temporal clusters of human cases of influenza A(H7N9) virus infection at county level in Zhejiang, Shanghai and Jiangsu provinces, China, 19 February?31 May 2013 (n=106)

________
Discussion
The outbreak of human infection with influenza A(H7N9) virus has attracted considerable attention because it is an emerging infectious disease caused by a novel, reassortant avian-origin influenza virus [26,27]. As this virus does not have the ability to be transmitted from person to person, it is unlikely to lead to a pandemic, but gene sequences analysis has indicated that it is better adapted than other avian influenza viruses to infect mammals [9,28]. Scientists are therefore concerned about the possibility of a pandemic if there is further gene mutation [29,30].
In the study period, the outbreak caused 131 confirmed human infections in mainland China. As reported previously [2], most cases were male, mainly aged 60 years or older. In contrast, the influenza A(H5N1) virus mainly affected children, adolescents and young adults (aged between 4 and 29 years, with a median of 18 years) [31]. As of 31 May, 38 cases of influenza A(H7N9) virus infection had died, with accumulative mortality of 29%, much higher than the 21% (17/82) in a previous report on the outbreak [2]. In comparison with the influenza A(H1N1)pdm2009 virus infections (mortality: 2.5%), the mortality of cases of influenza A(H7N9) virus infection was higher[32], although it was clearly lower than that of cases of highly pathogenic avian influenza A(H5N1) virus infection (mortality: 56%) [31]. As shown in a former case study [2], most of the human cases of influenza A(H7N9) virus infection had a severe illness (77/82), while others manifested as mild illness (4/82). In addition, a four year-old child in Beijing was identified as an asymptomatic carrier of influenza A(H7N9) virus [33]. This has been the only asymptomatic infection of influenza A(H7N9) virus to date. Thus, human infections with influenza A(H7N9) virus may result in fatal disease, mild illness as well as silent infection.
Our study shows that the influenza A(H7N9) outbreak in mainland China can be divided into 3 stages: early (weeks1?4), high-incidence (weeks5?9) and late stationary (weeks10?15). In the early stage, the outbreak was considered as non-threatening and highly sporadic. However, the high-incidence stage (in March?April) represented a peak, with 76% (100/131) of the cases during this stage. Furthermore, the three spatio-temporal clusters of cases identified were all during this period. Increased surveillance and control should be carried out in this period in future, to find out if there is an association with climate. In the late stationary stage, transmission was effectively controlled, leading to the ending of the outbreak in the most severely affected areas. This is now believed to be mainly due to the closure of live poultry markets [5].
Some studies have claimed that climatic factors, particularly temperature, have a clear impact on seasonal influenza outbreaks [34,35]. The weather in eastern China is very cold between December and January, and temperatures begin to rise in late February. It is usually relatively warm in March and April, and starts to get hot from May. The temporal characteristics of human infections with influenza A(H7N9) virus suggest that temperature has some association with incidence of this disease. The prevailing mild climate may have been particularly suitable for influenza A(H7N9) virus infection since there was an increase in the number of cases in March?April. If there is a relationship, attention must be paid as there could be a potential outbreak in the same period (March?April) in the future.
The outbreak was limited to 10 provinces in eastern China. In-depth studies of the factors driving the spatial diffusion of cases are essential to prevent future outbreaks. It is notable that when the first human infections appeared in Beijing and Henan, there were no cases in the neighbouring provinces. What is the reason for this? Were the two regions new, natural foci of the influenza A(H7N9) virus strain? Alternatively, and perhaps more probable, could they be attributed to the regional poultry trade or migration of infected wild birds? These possibilities need to be further studied.
The three coastal provinces of Zhejiang, Shanghai and Jiangsu were the most severely affected areas. Understanding the main factors causing this spatial concentration is critical in the approach to control this type of emerging disease. Some studies report that the spatial distribution of human infection with highly pathogenic avian influenza A(H5N1) is mainly associated with human population density, poultry density, elevation and the proportion of land covered by water bodies [16,36]. However, our study suggests that population density does not influence the spatial pattern of human infection with influenza A(H7N9) virus. Further studies are needed to confirm whether or not there are other counterintuitive correlations involved in the spatial distribution of human infection due to this virus.
The SatScan method has become an increasingly popular adjunct for exploring the spatio-temporal distribution of infectious diseases [37]. We selected this method in the hope of identifying clustering of cases and found three clusters in the three coastal provinces with a high incidence, which were proposed to be the most probable place of the viral reassortment [12,38]. Two of the identified clusters represent the most important hotspots of the outbreak in China and were found in the counties along the boundaries between these three provinces. Surveillance should be focused on these regions to identify any future emergence of this or other new virus strains and monitor possible further mutation or reassortment that may alter the risks to animals or humans. The third cluster covered just two counties within Jiangsu province and was situated far from the other two clusters. In addition, its duration was very short. The risk factors that contribute to clustering warrant further study.
This study, using spatio-temporal methods in conjunction with classic descriptive analysis, found evidence of clustering, in both space and time. The results can provide clues for further developments and serve as a reference and basis for the surveillance and control of human infection with influenza A(H7N9) virus in the future.
_____
Acknowledgements
This work was supported by the Jiangsu Province Health Development Project with Science and Education (ZX201109, RC2011085 and RC 2011094), and the research project of Jiangsu preventive medicine (NO.YZ201020).
Conflict of interest
None declared.
Authors? contributions
Wendong Liu and Kun Yang contributed equally to this study. Correspondence to Yefei Zhu and Fenyang Tang. Yefei Zhu and Fenyang Tang designed the study; Wendong Liu, Kun Yang, Xian Qi, KeXu, Hong Ji, Jing Ai, Aihua Ge, Ying Wu, Yuan Li, Qigang Dai, Qi Liang and Changjun Bao collected, analyzed and interpreted data; Wendong Liu, Kun Yang, and Yefei Zhu drafted the article; Robert Bergquist revised the manuscript. All authors reviewed and revised the first and final drafts of this manuscript.
References
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W,et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888-97. http://dx.doi.org/10.1056/NEJMoa1304459 PMid:23577628
- Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H,et al. Preliminary report: epidemiology of the avian influenza A (H7N9) outbreak in China. N Engl J Med. 2013 Apr 24. [Epub ahead of print]. http://dx.doi.org/10.1056/NEJMoa1304617 PMCid:PMC3652256
- Alcorn T. As H7N9 spread in China, experts watch and wait. Lancet. 2013;381(9875):1347. http://dx.doi.org/10.1016/S0140-6736(13)60868-5
- Hvistendahl M. Influenza. A decade after SARS, China's flu response wins cautious praise. Science. 2013;340(6129):130. http://dx.doi.org/10.1126/science.340.6129.130 PMid:23580499
- The Lancet Infectious Diseases. A proportionate response to H7N9. Lancet Infect Dis.2013;13(6):465. http://dx.doi.org/10.1016/S1473-3099(13)70134-8
- Alcorn T. China's H7N9 outbreak slows but experts remain wary. Lancet Respir Med. 2013;1(4):286. http://dx.doi.org/10.1016/S2213-2600(13)70092-4
- Ai J, Huang Y, Xu K, Ren D, Qi X, Ji H, et al. Case-control study of risk factors for human infection with influenza A(H7N9) virus in Jiangsu Province, China, 2013. Euro Surveill. 2013;18(26):pii=20510.Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20510 PMid:23827526
- Baas C, Barr IG, Fouchier RA, Kelso A, Hurt AC. A comparison of rapid point-of-care tests for the detection of avian influenza A(H7N9) virus, 2013. Euro Surveill. 2013;18(21):pii=20487.Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20487
- Chen Y, Liang W, Yang S, Wu N, Gao H, Sheng J, et al. Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet. 2013;381(9881):1916-25. http://dx.doi.org/10.1016/S0140-6736(13)60903-4
- Han J, Niu F, Jin M, Wang L, Liu J, Zhang P, et al. Clinical presentation and sequence analyses of HA and NA antigens of the novel H7N9 viruses. Emerg Microbes Infect. 2013;2:e23. http://dx.doi.org/10.1038/emi.2013.28 PMCid:PMC3675404
- Hu Y, Lu S, Song Z, Wang W, Hao P, Li J, et al. Association between adverse clinical outcome in human disease caused by novel influenza A H7N9 virus and sustained viral shedding and emergence of antiviral resistance. Lancet. 2013;381(9885):2273-9. http://dx.doi.org/10.1016/S0140-6736(13)61125-3
- Liu D, Shi W, Shi Y, Wang D, Xiao H, Li W, et al. Origin and diversity of novel avian influenza A H7N9 viruses causing human infection: phylogenetic, structural, and coalescent analyses. Lancet. 2013;381(9881):1926-32. http://dx.doi.org/10.1016/S0140-6736(13)60938-1
- Cohen J. Influenza. New flu virus in China worries and confuses. Science. 2013;340(6129):129-30. http://dx.doi.org/10.1126/science.340.6129.129 PMid:23580498
- Meng B, Wang J, Liu J, Wu J, Zhong E. Understanding the spatial diffusion process of severe acute respiratory syndrome in Beijing. Public Health. 2005;119(12):1080-7. http://dx.doi.org/10.1016/j.puhe.2005.02.003 PMid:16214187
- Lai PC, Kwong KH, Wong HT. Spatio-temporal and stochastic modelling of the severe acute respiratory syndrome (SARS).Geospat Health. 2013. Forthcoming.
- Martin V, Pfeiffer DU, Zhou X, Xiao X, Prosser DJ, Guo F, et al. Spatial distribution and risk factors of highly pathogenic avian influenza (HPAI) H5N1 in China. PLoSPathog. 2011;7(3):e1001308. http://dx.doi.org/10.1371/journal.ppat.1001308 PMid:21408202 PMCid:PMC3048366
- Lee SS, Wong NS. The clustering and transmission dynamics of pandemic influenza A (H1N1) 2009 cases in Hong Kong. J Infect. 2011;63(4):274-80. http://dx.doi.org/10.1016/j.jinf.2011.03.011 PMid:21601284
- National Health and Family Planning Commission of the People's Republic of China (NHFPC).Diagnostic and treatment protocol for human infections withavian influenza A (H7N9) (2nd edition, 2013). Beijing: NHFPC; 11 Apr 2013.Available from: http://www.moh.gov.cn/yjb/bmdt/201304/9e989eba0d0d4500ba5dbb89c3bd7829.shtml
- Chinese Center for Disease Control and Prevention (CDC). [Sex-age specific population of each county in mainland China in 2011]. Beijing: Chinese CDC. Scientific Data Sharing Center of Public Health. [Accessed 20 June 2013]. Chinese. Available from: http://www.phsciencedata.cn/Share/ky_sjml.jsp?id=454aaff4-a4ba-41e3-accc-fefc06900a43
- National GeomaticsCenter of China (NGCC).[Maps].Beijing: NGCC. [Accessed 15 June 2013]. Chinese. Available from: http://ngcc.sbsm.gov.cn/article/sjcg/dtxz/
- Lai PC, So FM, Chan KW. Spatial epidemiological approaches in disease mapping and analysis. Boca Raton, FL: CRC Press; 2008.
- Cuzick J, Edwards R. Spatial clustering for inhomogeneous populations. J R Stat Soc Series B Stat Methodol. 1990;52:73-104.
- Kuldorff M, Information Management Services, Inc. SaTScan v9.0: software for the spatial and space-time scan statistics. Boston, MA: SaTScan; 2010. [Accessed 10 Mar 2011].Available from: http://www.satscan.org/download.html
- Kulldorff M, Nagarwalla N. Spatial disease clusters: detection and inference. Stat Med. 1995;14(8):799-810. http://dx.doi.org/10.1002/sim.4780140809
- Kulldorff M. A spatial scan statistic. Commun Stat Theory Methods. 1997;26(6):1481-96. http://dx.doi.org/10.1080/03610929708831995
- Horby P. H7N9 is a virus worth worrying about. Nature. 2013;496(7446):399. http://dx.doi.org/10.1038/496399a PMid:23619655
- Uyeki TM, Cox NJ. Global concerns regarding novel influenza A (H7N9) virus infections. N Engl J Med. 2013;368(20):1862-4. http://dx.doi.org/10.1056/NEJMp1304661 PMid:23577629
- Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, et al. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature.2013;501(7468):556-9. http://dx.doi.org/10.1038/nature12391 PMid:23842497
- Tharakaraman K, Jayaraman A, Raman R, Viswanathan K, Stebbins NW, Johnson D,et al. Glycan receptor binding of the Influenza A Virus H7N9 hemagglutinin. Cell. 2013;153(7):1486-93. http://dx.doi.org/10.1016/j.cell.2013.05.034 PMid:23746830
- Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y,et al. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18(15):pii=20453.Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20453 PMid:23594575
- Fiebig L, Soyka J, Buda S, Buchholz U, Dehnert M, Haas W. Avian influenza A (H5N1) in humans: new insights from a line list of World Health Organization confirmed cases, September 2006 to August 2010. Euro Surveill. 2011;16(32):pii=19941.Available from: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19941 PMid:21871222
- Yu H, Feng Z, Uyeki TM, Liao Q, Zhou L, Feng L, et al. Risk factors for severe illness with 2009 pandemic influenza A (H1N1) virus infection in China. Clin Infect Dis. 2011;52(4):457-65. http://dx.doi.org/10.1093/cid/ciq144 PMid:21220768 PMCid:PMC3060897
- Butler D. H7N9 bird flu poised to spread. Nature. 15 April 2013.
- Lowen AC, Mubareka S, Steel J, Palese P. Influenza virus transmission is dependent on relative humidity and temperature. PLoS Pathog. 2007;Oct 19;3(10):1470-6. http://dx.doi.org/10.1371/journal.ppat.0030151 PMid:17953482 PMCid:PMC2034399
- Tamerius J, Nelson MI, Zhou SZ, Viboud C, Miller MA, Alonso WJ. Global influenza seasonality: reconciling patterns across temperate and tropical regions. Environ Health Perspect. 2011;119(4):439-45. http://dx.doi.org/10.1289/ehp.1002383 PMid:21097384 PMCid:PMC3080923
- Kuo HI, Lu CL, Tseng WC, Li HA. A spatiotemporal statistical model of the risk factors of human cases of H5N1 avian influenza in South-east Asian countries and China. Public Health, 2009;123(2):188-93. http://dx.doi.org/10.1016/j.puhe.2008.10.012 PMid:19144364
- Kulldorff M, Heffernan R, Hartman J, Assun??o R, Mostashari F. A space?time permutation scan statistic for disease outbreak detection. PLoS Med. 2005;2(3):e59. http://dx.doi.org/10.1371/journal.pmed.0020059 PMid:15719066 PMCid:PMC548793
- Xiong C, Zhang Z, Jiang Q, Chen Y. Evolutionary characteristics of A/Hangzhou/1/2013 and source of avian influenza virus H7N9 subtype in China. Clin Infec Dis.2013;57(4):622-4. http://dx.doi.org/10.1093/cid/cit294 PMid:23650290
-
--------