Re: 225G Preliminary Worldwide Tracking & Evaluation
from NS1 via email -
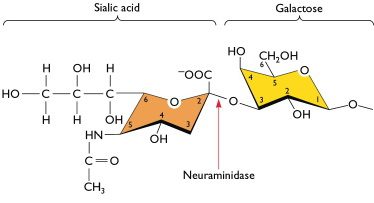
"We appreciate and agree with Dr. Racaniello?s astute analysis on 225G. We are slightly more cautious about assigning ?low? transmissibility based on the limited evidence we have, though the ferret studies and current lack of defined human clusters compel one toward an initial evaluation of low transmissibility. The absence of evidence is not always negational. More data points will define the matter soon."
Originally posted by Florida1
View Post
"We appreciate and agree with Dr. Racaniello?s astute analysis on 225G. We are slightly more cautious about assigning ?low? transmissibility based on the limited evidence we have, though the ferret studies and current lack of defined human clusters compel one toward an initial evaluation of low transmissibility. The absence of evidence is not always negational. More data points will define the matter soon."
Comment